Type IIS restriction endonucleases mediated cloning
Type IIS-mediated assembly methods are based on the original description of "Golden Gate" cloning (Engler et al. 2008) and allows efficient simultaneous assembly of multiple DNA parts, without requiring the isolation of individual fragments.
Nicola Patron has assembled a short online course on Type IIS that covers:
How type IIS restriction enzymes enable a one-pot reaction
How to easily and reliably assemble multiple standard parts in a single reaction, and
How to make new parts
Type IIS methods in plants
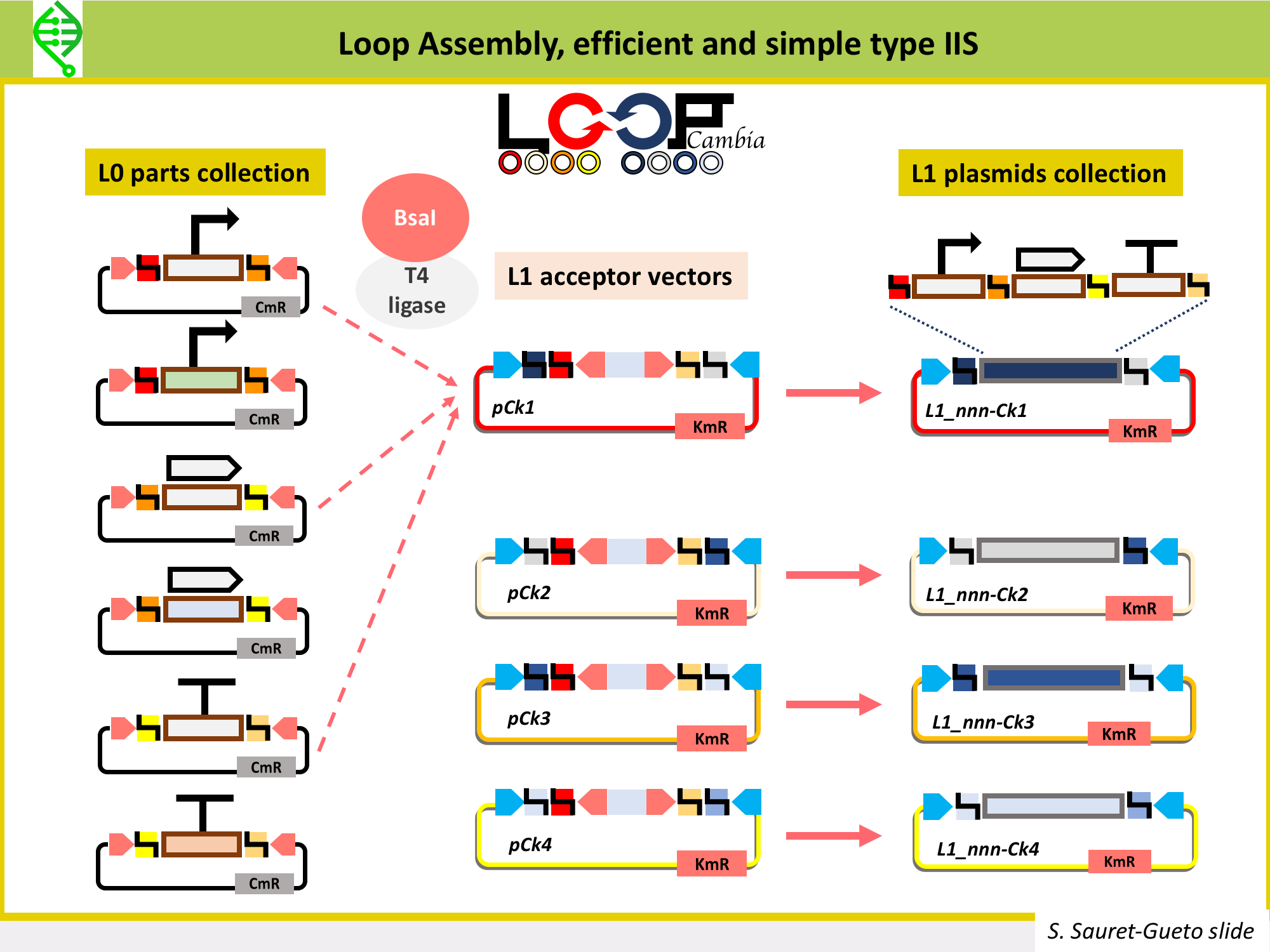
There are several type IIs methods being used to make constructs to transformplants, all of them use the same common syntax and fusion sites for L0 parts in order to make the building of L1 parts a standard process. These type IIS methods are: MoClo, GoldenBraid and Loop Assembly. All of them use the same typeIIS enzyme, BsaI, to assemble L0 parts into L1 parts. Thus, all L0 parts need to be domesticated for BsaI sites (so, all BsaI sites need to be removed from the sequence).
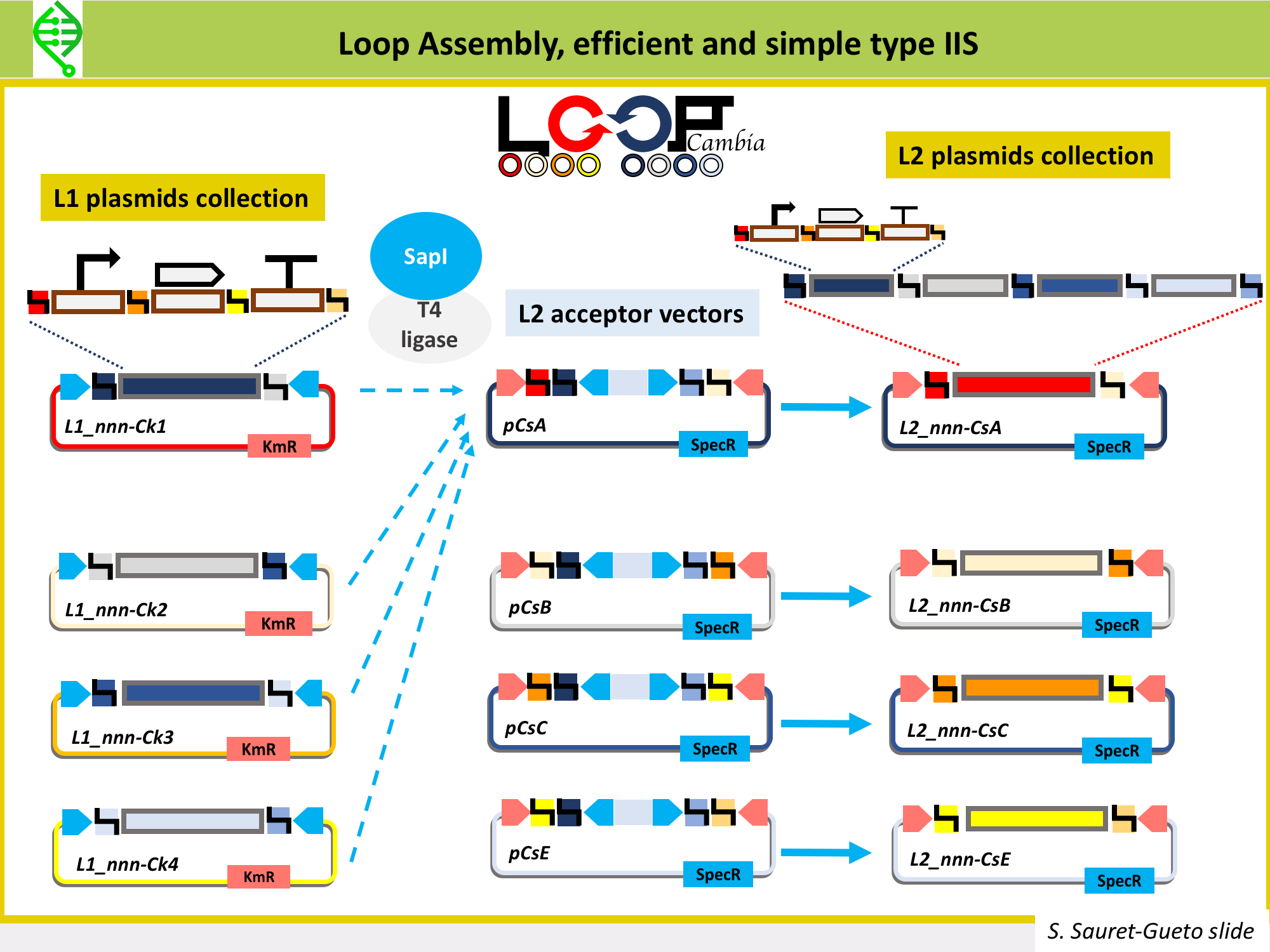
MoClo, GoldenBraid and Loop Assembly differ in the tyep IIS enzyme used to go from L1 into L2 vectors, as well as on number of plasmids needed to build the L1 and L2, the flexibility of moving in between levels, and the option of cloning plasmids under openMTA.
Loop Assembly has been developed in the Haseloff Lab by Bernardo Pollak and Fernarn Federici, is an efficient and simple type IIS method that uses BsaI and SapI as type IIS enzymes, and two sets of 4 plasmids for cloning from one level into the next. This is the method being used in Marchantia to assemble L1 and L2 constructs. Thus, the Marchantia L0 parts are domesticated for BsaI and SapI. More details on Loop here.